Biosensors
A core objective of our research program is to understand the mechanism of the transduction of chemical information at interfaces and to use this knowledge in the pursuit of innovative sensors. A sensor is a device that responds to physical, chemical or biological quantity, and converts that quantity into a signal or an output. An example of a sensing process (from molecular recognition to signal output) is shown in Figure 1. The driving force behind our biosensor research is to build sensor systems that quantitatively measure target species in a complex system.

We have also developed sensors for Microcystin toxins, cyanobacteria, organophosphates, endocrine-disrupting chemicals, oral proteins, nitrobenzenes, fullerenes, and heavy metals. Some of our earlier sensors have been used for the detection of trace uranium and vanadium.We reported the basic idea of a capillary biosensor which uses the waveguiding properties of the capillary to integrate the signal over an increased surface area without simultaneously increasing the background noise from the detector (“Integrating Waveguide Biosensor,” Analytical Chemistry, 74,713-719, 2002). We have demonstrated the application of the idea in immunochemical and nucleic acid recognition (Biosensor Bioelectronics, 18, 1135-1147, 2003).
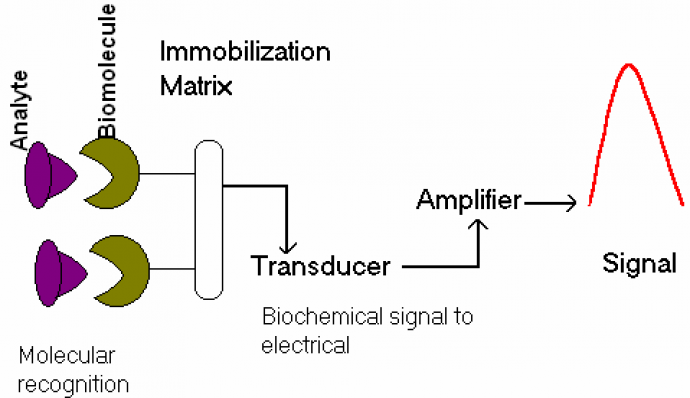
Figure 1: molecular recognition process at the sensor-analyte interface
Development of biosensors for detection of biomarkers of chronic back pain: Chronic Low Back Pain (cLBP) is a cause for concern as it has the potential to significantly affect function, thus limiting the activities of daily living resulting in socio-economic issues. Back pain is an exceedingly common condition affecting 70–85% of all people during their lifetimes. Our group has reported the first electrochemical characterization of pain biomarkers which include Arachidonic acid (AA), Prostaglandin G2 (PGG2), and Cyclooxygenase II (COX-2).
We have also developed an electrochemical immunosensor for monitoring these pain biomarkers. Our results revealed that direct electron transfer between arachidonic acid metabolites and the electrode could be easily controlled and that an enzyme-modified electrode dramatically enhanced bio-electrocatalytic activity towards AA. Cyclic voltammetric analysis of AA revealed a concentration-dependent anodic current with a slope of 2.37 and a limit of detection of 0.25 nM.
This unique AA/Au-electrode electron transfer provides a good electrochemical-sensing platform for PGH2 as the basis for quantitation of pain. The BioSMART Pain team had studied COX-2 and inducible Nitrous oxide synthase (NOS) as potential biomarker ability of serum COX-2 and iNOS as an objective measure of pain level for ~ 800 Turkish and American patients. The sensors we developed have the potential to provide an objective characterization of pain biomarkers for clinical diagnoses.
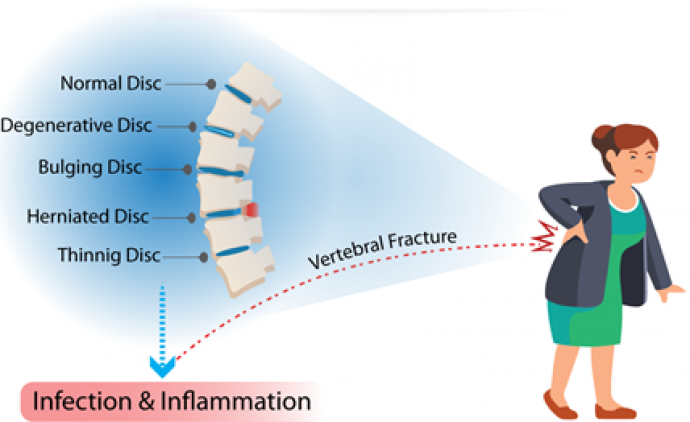
Figure 2: Biosensors for chronic lower back pain
Handheld multiarray biosensors for surveillance and rapid detection: Catastrophic plant diseases worsen the global food supply thereby affecting millions of people worldwide. Plant pathogens are difficult to control because their populations are variable in time, space, and genotype. Most insidiously, they evolve, often overcoming resistance and evading the attention of farmers. Funded by Bill and Melinda Gates Foundation and the NSF, we have developed sensors for the blue-green mold Penicillium italicum, which is among the most problematic post-harvest plant infections limiting the integrity of citrus and many other crops during storage and transportation. A multi 96-well plate gave the most sensitive detection with a 4 x 102 spores/mL limit of detection; a linear dynamic range between 2.9 x103 - 6.02 x104 spores/mL (R2 0.9939) and standard deviation of less than 5% for five replicate measurements. The selectivity of the ligands was tested against Trichaptum biforme, Glomerulla cingulata (Colletotrichum gloeosporioides) and Aspergillus nidulans fungi species (#3).
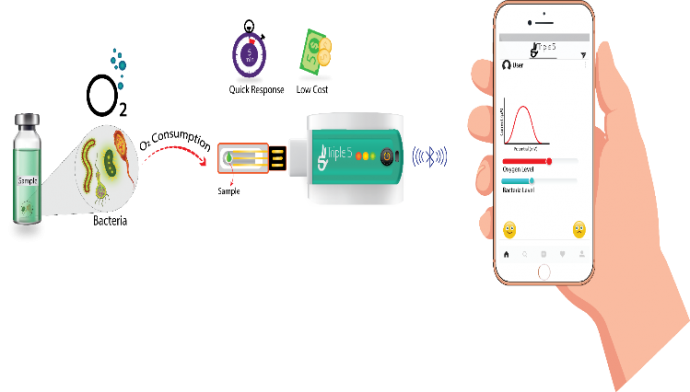

Figure 3: Design concept (left figure) and prototype for the hand-held sensor (right figure)
Automated microbial cell culture, sensing, and classification system (AMC3) is an integrated sensor array platform that was developed in our laboratory. The system is composed of a multi-potentiostat, an automated data collection system (Python program, Yocto Maxi-coupler electromechanical relay module), and a powerful classification program. The classification scheme consists of Probabilistic Neural Network (PNN), Support Vector Machines (SVM), and General Regression Neural Network (GRNN) oracle-based system. Differential Pulse Voltammetry (DPV) is performed on standard samples or unknown samples.
Then, using preset feature extractions and quality control, accepted data are analyzed by the intelligent classification system. In typical use, thirty-two extracted features were analyzed to correctly classify the following pathogens: Escherichia coli ATCC#25922, Escherichia coli ATCC#25922, Escherichia coli ATCC#11775, and Staphylococcus epidermidis ATCC#12228. 85.4% accuracy range was recorded for unknown samples, and within a shorter period than the industry standard of 24 hours (#15, 34-36, 52, 74).
