Metabolomics Shared Resource of Rutgers Cancer Institute of New Jersey
Services, Technologies, Instrumentation
Training
The Metabolomics shared resource offers seminars and presentations by internal and external experts. In addition, it trains users in the Metabolomics Analysis and Visualization Engine (MAVEN) software package developed in the Rabinowitz lab (Anal. Chem. 2010, 82, 9818–9826) for data processing and visualization. To facilitate the training of graduate students and postdocs, professional staff manually review the LC-MS raw data of less experienced users, to ensure accurate processing and interpretation. Facility staff members also routinely manually check all data associated with key biological discoveries.
Expert Consultation
Investigators planning to conduct research using the Metabolomics shared resource are encouraged to submit an experimental plan describing the biological question and protocol design. Users are guided to detailed SOPs for critical steps like sample harvesting and metabolite extraction. Given the unique requirements of many projects, shared resource personnel are available to meet with investigators at the inception of the project. Consultation regarding experimental design is also routinely provided prior to grant submission. After sample analysis, experimental data is provided to the investigator along with suggestions for interpretation. Additional experiments may be planned with the investigator to optimize the results.
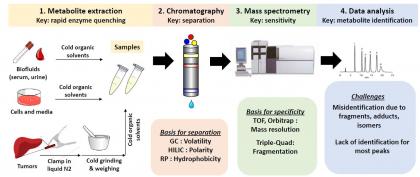
Figure 1. Metabolomics Workflow
Water-Soluble Metabolite Quantitation
The identification and quantitation of polar metabolites by LC-MS (Fig. 1) is our most popular service. Most classical intermediary metabolites are polar, including glycolytic intermediates, TCA compounds, amino acids, nucleotides, and derivatives. Specificity is achieved through the combination of chromatographic separation and high-resolution MS with MS/MS used in selected cases to differentiate isomers. Current metabolomics methods cover approximately 300 water-soluble metabolites, each identified by retention time match to synthetic standards. In addition, we offer dedicated methods for analytically challenging subclasses of metabolites such as folates (Anal Bioanal Chem 2017, 409, 5955-5964). Data quality is the top priority with methods validated for analyte stability, detection specificity, and quantitative reproducibility (Annu Rev Biochem, 2010, 86, 277-304). For researchers primarily interested in central carbon metabolism, it is common to analyze only anionic metabolites (carboxylic acids and phosphorylated compounds), which can be achieved with a single LC-MS run in negative ion mode. To also cover cationic metabolites like acyl-carnitines, nucleosides, and polyamines, users may elect to run in positive ion mode. Stable isotope labeling patterns can also be routinely measured (13C, 15N, 2H, etc.). Certain important metabolites like glycerol and acetate are not measured well by LC-MS; newly developed methods enable their measurement by GC-MS.
Lipid Quantitation
Identification and quantitation of non-polar metabolites by LC-MS. Lipidomics methods enable measurements of the concentrations of approximately 500 lipids spanning all major classes. For isotope tracer studies, due to the challenge in extracting labeling data across such a complex mixture of lipids, users typically focus on saponified fatty acids. The labeling pattern of fatty acids provides high-value information regarding the metabolic sources of their building blocks: acetyl-CoA and NADPH (J Am Chem Soc 2017, 139, 14368-14371, ; Cancer Metab 2014).
Novel or Unexpected Metabolite Discovery (additional data analysis at no cost)
The analytical methods used for metabolite quantitation are untargeted: They measure all ions present in the sample. From these measurements, we routinely quantitate only known analytes, because most users are focused on the abundances and labeling of these classical species (like glutamine, ATP, fructose bisphosphate), and do not wish to be distracted by the thousands of other unidentified peaks, most of which reflect environmental contamination or mass spectrometry artifacts, not new metabolites (Anal Chem 2017). Nevertheless, the discovery of new cancer-associated metabolites is a great scientific opportunity, born out by the clinical importance of 2-hydroxyglutarate (Nature 2010). The MAVEN software, as well as online tools like XCMS (Anal Chem 2006), enable users to find unexpected peaks that go up or down in a particular set of samples. The shared resource then works with users to identify the underlying metabolites. To this end, the core is developing novel strategies for metabolite identification based on combining isotope labeling and MS/MS.
Glucose Uptake, Lactate Uptake, and Oxygen Consumption
To measure glucose and glutamine uptake and lactate and glutamate excretion, the Metabolomics shared resource has YSI/NOVA electrochemical analyzers at both the Princeton and New Brunswick sites. For kinetic measurements of oxygen uptake and lactate excretion, Seahorse Bioscience XF photochemical flux analyzers are located at both sites.
Instrumentation
The shared resource has sites in both Princeton and New Brunswick. The Princeton site is located in ~1500 sq. ft. on the second floor of the Frick Chemistry Building. The New Brunswick site is located in ~800 sq. ft. on the second floor of CINJ and room 4253 of the Rutgers Child Health Institute. Instrumentation includes:
- Thermo TSQ Quantum Discovery Max triple quadrupole mass spectrometer coupled with Shimadzu HPLC system (Princeton, 2006)
- Thermo Exactive mass spectrometer coupled with Accela UPLC system (Princeton, 2008)
- Seahorse XF Extracellular Flux Analyzer (Princeton, 2008)
- Seahorse XF Extracellular Flux Analyzer (New Brunswick, 2012)
- NOVA Electrochemical Analyzer (New Brunswick, 2012)
- Agilent 6550 iFunnel QTOF mass spectrometer coupled with Agilent Infinity 1290 UHPLC system (Princeton, 2013)
- Thermo Q-Exactive MS/MS coupled with Thermo UPLC system (Princeton, 2015)
- YSI Automated Electrochemical Analyzer (Princeton, 2016)
- Thermo Q Exactive MS/MS coupled with Thermo UPLC system (New Brunswick, 2017)
- Bruker SolariX XR high-res imaging mass spectrometer (or similar, purchase pending at Princeton)
